Realize the scientific potential of single-cell analysis
Single-cell analysis technologies have facilitated significant developments and discoveries in translational biology. Continued growth and transformation of the tools surrounding the single-cell workflow are required to meet the high bar for quality data produced and quantity of useable data per experiment.
Scientists often discover these issues too late in the experimental pipeline, which contributes to increasing frustration and unanticipated costs in irreplaceable lost samples, time, resources, and funds. Single-cell analysis technologies have facilitated significant developments and discoveries in translational biology.
Enable samples to tell their native truth by maximizing the data obtained from each and every cell analyzed.

Sample preparation methods highly influence the quality of sample input into single-cell sequencing techniques. Traditional methods can damage cells, cause gene expression changes or simply cannot deliver the sample quality or quantity – resulting in inaccurate population representation of the biological sample or insufficient cells for a complete experiment.
The transformative potential of Levitation Technology™ and the LeviCell® systems for multimodal single-cell analysis is clear
LevitasBio® has developed an innovative Levitation Technology that uses less than 1 psi of pressure to enable a touch-free, label-free, three- step enrichment process that takes only 20 minutes to complete. This can be applied seamlessly to a wide range of research areas and workflows.
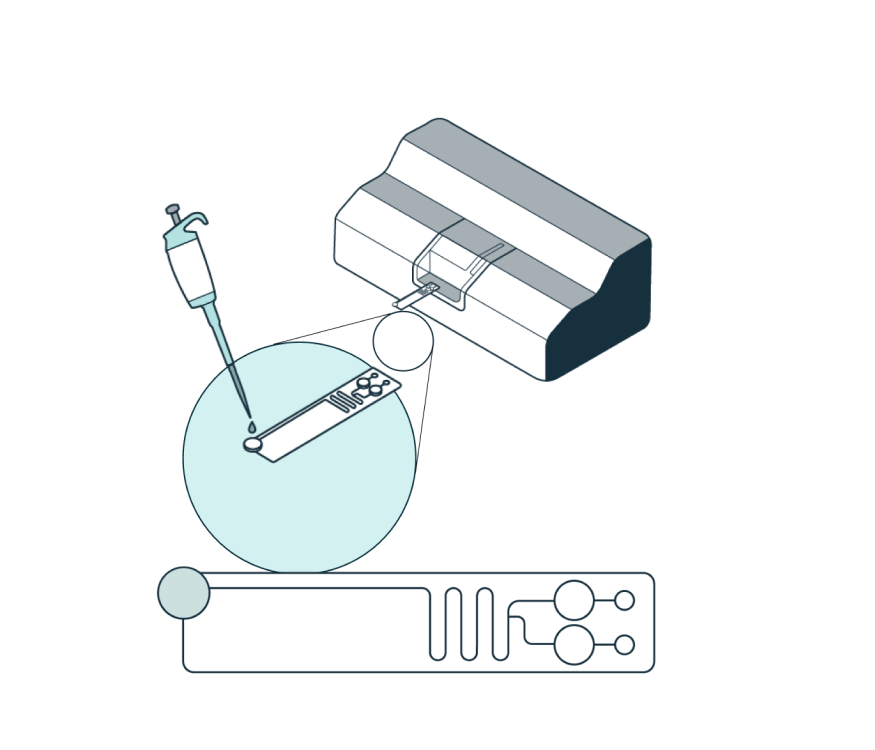
Prepare the sample by adding the Levitation Agent, a paramagnetic solution. After inserting the cartridge into the system, dispense the sample into the inlet well.

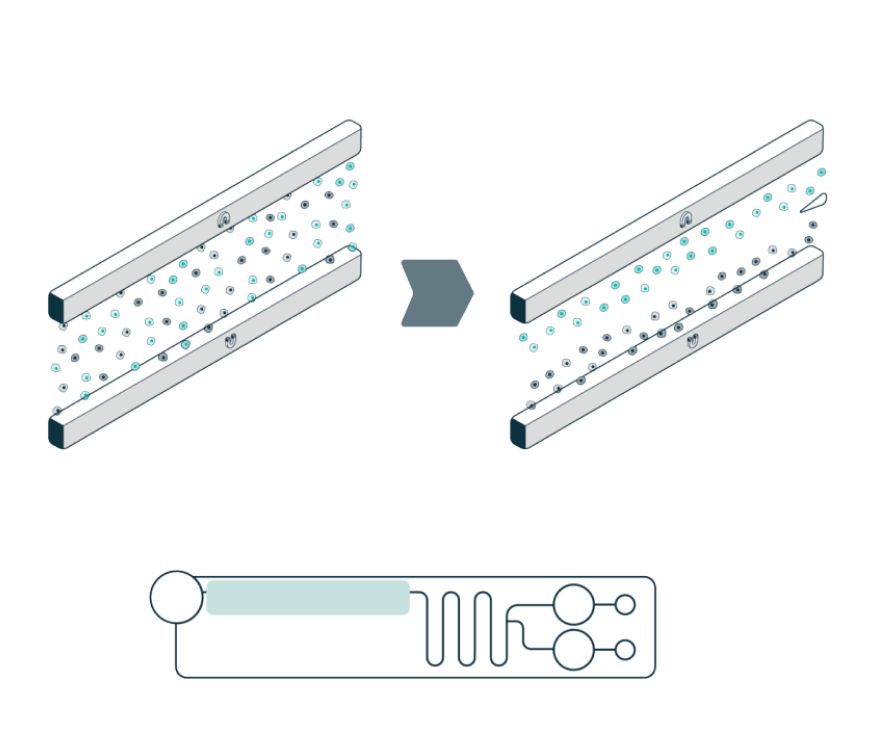
The system automatically loads the sample into the separation channel while the sample is imaged and levitated. The system performs automated, label-free enrichment plus debris removal via levitation.

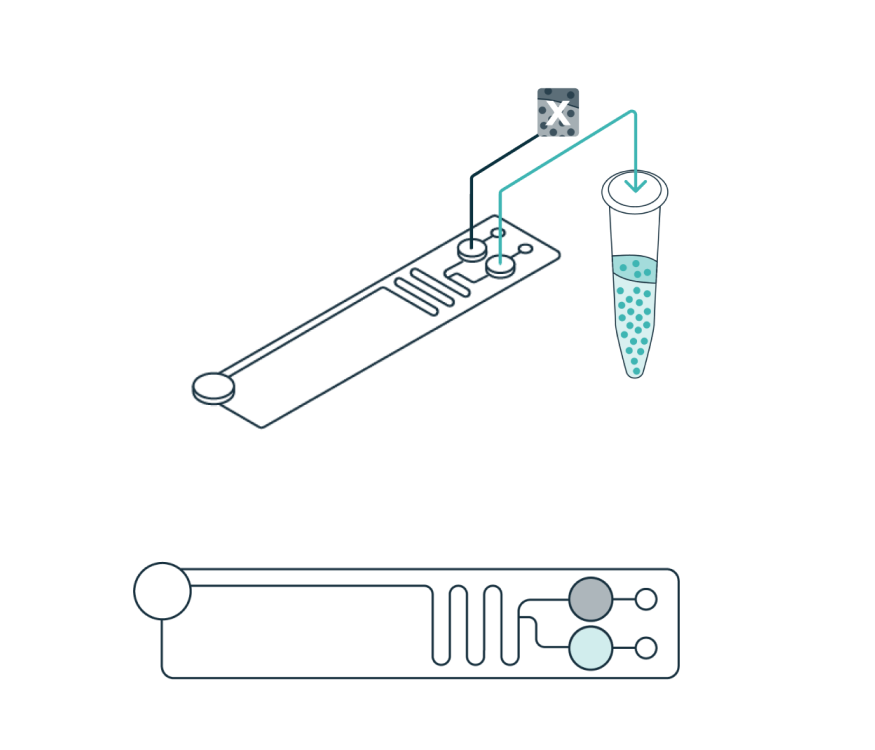
Once the cells have levitated to their final equilibrium position, a live view in the software enables the user to define how the sample will be divided (fraction of interest and waste). Separated fractions are collected into the cartridge outlet wells.

Viable cells levitate much higher than dead cells and debris, allowing for clear separation and collection by the LeviCell systems. Start with high-quality cells every time for the best and most accurate results. This gentle yet effective process makes it possible.
Maximize cell viability
and enrichment
Preserve of cell
population representation
Minimize the risk of
contamination and damage
Maximize data for
every sample analyzed
The innovative label-free separation technology of the LeviCell systems facilitate complete debris and dead cell removal without affecting the original population representation of gene expression (please see our webinar summary)
When it comes to single-cell study, purity and viability of cells harvested from the sample preparation stage is a determining factor for generating high quality data in downstream assays such as NGS (Next Generation Sequencing).
The 10x Genomics single-cell sequencing platforms have helped to revolutionize our understanding of biology and how the measurement of individual cells’ gene expression can uncover the previously hidden messages commonly unseen in bulk mRNA experiments.
Unfortunately while the single cell sequencing workflow has proven to be fast, simple and reliable, obtaining the most complete and accurate single cell data depends on up-front sample preparation steps. Traditional methods currently in use tend to damage cells, have low capture efficiency, and often completely fail to process sensitive and difficult to work with samples such as dissociated tissue samples. The most common sample enrichment methods, flow cytometry and bead-based enrichment methods, both often lead to significant cell stress and gene expression changes due to the high pressures used (Xiong et al., 2002; Romero-santacreu et al., 2009; Van Den Brink et al., 2017) or cell signaling effects due to the cell surface binding required by these methods (Kornbluth and Hoover, 1989; Christaki et al., 2011). Due to these shortcomings many promising single cell experiments delivered biased results or fail to delivery results entirely.
The urgency for technological platforms that are gentle (to minimize cell damage), and efficient (to maximize the capture of all live cells in the population while maintaining the original cellular representation) is imperative to realize the scientific and clinical potential of single-cell analysis.
The LeviCell systems demonstrates such a promise.
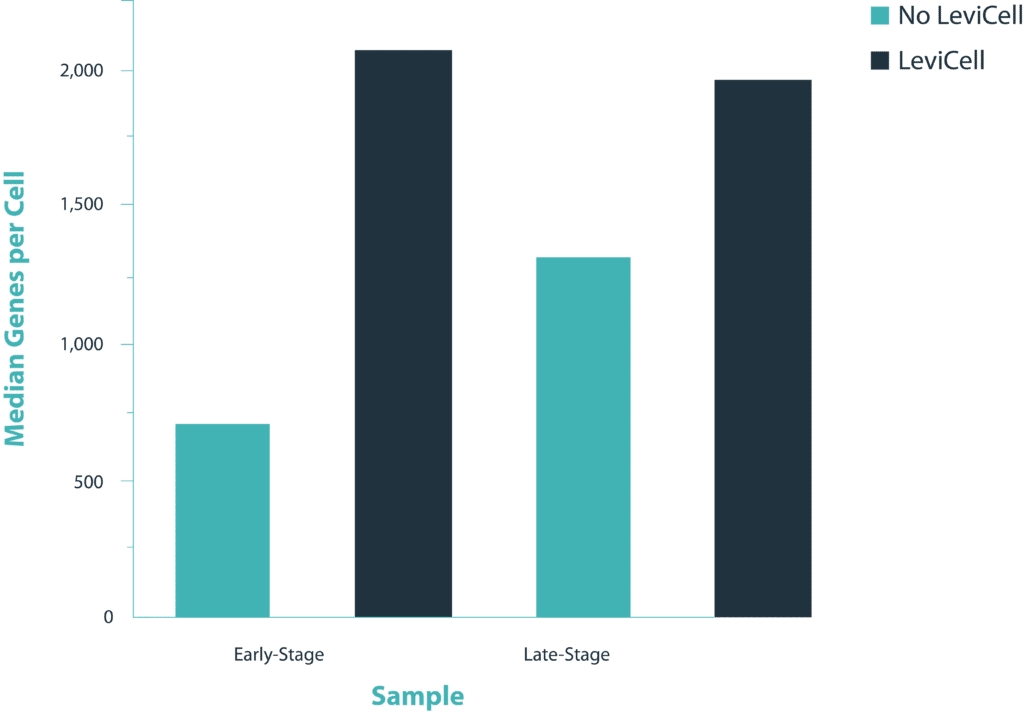
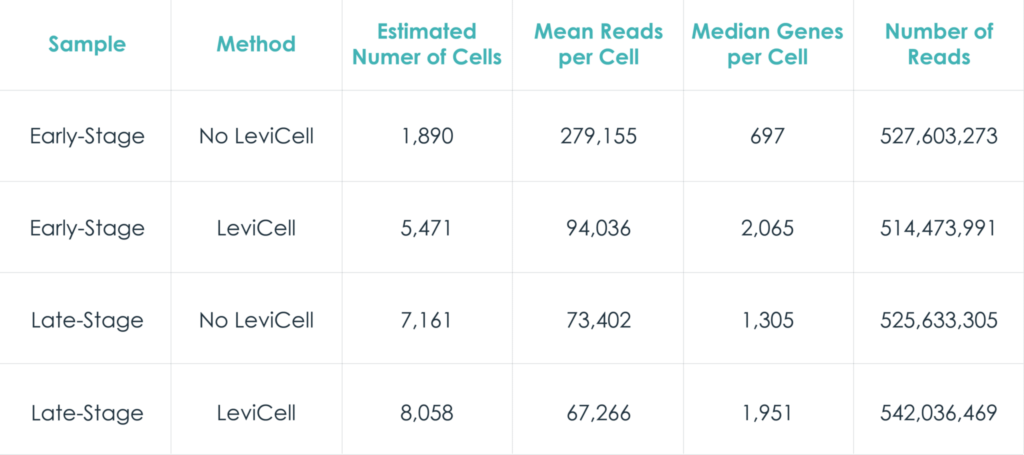
To demonstrate the benefits of the LeviCell systems in removing dead cells and debris, each sample of 400k cells was split into fractions prior to the each enrichment process. One half of each sample was washed into a 10x library preparation buffer, while the other half was enriched for viable cells using the LeviCell 1.0 system . After quantitation, the output from the LeviCell 1.0 was put directly into the Chromium Next GEM single Cell 3’ kit step without additional washing. Approximately 15k cells were used for each method. Final cDNA concentration was estimated by BioAnalyzer. While all samples succeeded at this step, there was a clear improvement in quality of the cDNA and the results library quality from the material generated by the LeviCell.
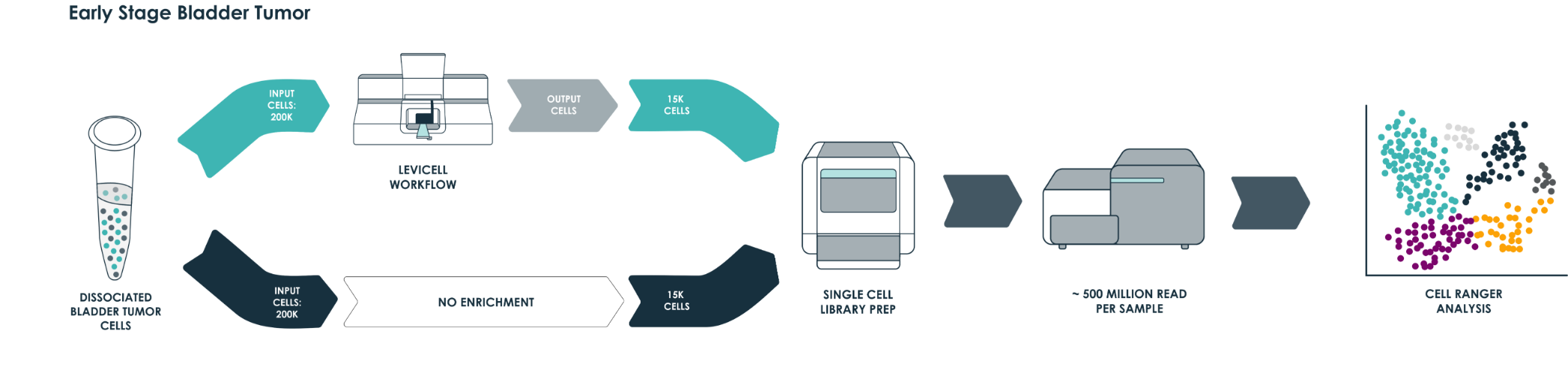
All samples were normalized before library preparation to account for differences yields, after normalization 15K cells were loaded from each process into the Chromium system.
The data on the right show how LeviCell’s gentle cell separation approach enables the study of notoriously delicate and sensitive primary cell types without compromising the integrity of the original cell populations. Sorted and unsorted ovarian DTC samples were blocked with TruStain human Fc Block for 15 minutes at RT then stained with 5 μL each of anti-CD45, anti-CD19, anti-CD11b, and 10 μL of anti-CD3, for 30 minutes at room temperature. Samples were washed with FACS buffer (0.5% BSA in PBS) once before analyzing on a Sony SH800S cytometer. The LeviCell output shows a similar amount of CD3+, CD19+, and CD11b+ cells within the CD45+ lymphocyte population while the sample processed by the bead method shows reduced proportions of CD3+ and CD11b+ cells.
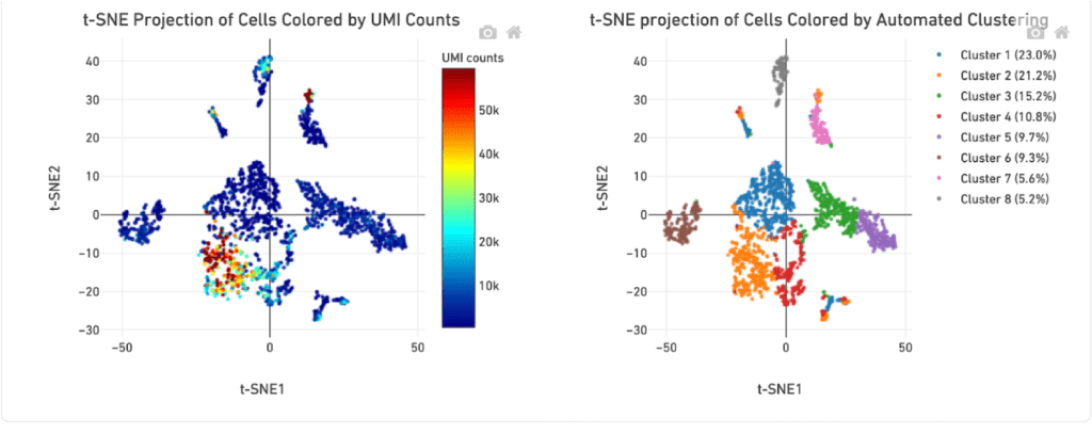
The distribution of UMI counts per cell in the standard method shows a small cluster of cells with the majority of the UMI counts. This same cluster of cells is represented by a single cluster when grouped by gene expression changes. In contrast, in the LeviCell enrichment sample there is a larger number of cells with significant numbers of UMI counts. This same group of cells is represented by four gene expression clusters (lower right), indicating a larger range of cellular diversity captured in the sequencing results.
A similar trend is observed with the late stage cancer sample, although the trends are less pronounced. Here, the sample processing with the standard method shows a large number of cells with high diversity of UMI counts, and correspondingly five clusters of gene expression. After enrichment on the LeviCell 1.0 system, the sample has a larger number of cells with high diversity of UMI counts, and six clusters of gene expression clusters, indicating a superior view of the expression profiles.

The LeviCell systems ability to seamlessly enrich for target cells and produce robust yields of viable cells without preferentially depleting or changing the frequency or expression of cell types gives it the necessary technological characteristics that many scientists have been waiting for to take them to the next level.