As a Senior Scientist at the NCI Cancer Genomics Research Lab, Seth Brodie was one of the first people to work with the beta version of the LeviCell system. He was so impressed by the technology’s potential that he took a position with LevitasBio and now serves as Associate Director of Research and Development where he focuses on applications development surrounding single-cell genomics.

We had a chance to sit down recently and chat with Dr. Brodie about his early experiences with the pre-commercial and commercial versions of the LeviCell. Below are highlights of our conversation.
Answers have been edited for clarity.
How did you first become involved in testing the LeviCell? What sample types were you working with and what types of questions were you investigating?
We were one of the first instrument placements. This was the pre-commercial instrument that worked on the same principle, but it was a vastly different instrument. It was a permanent flow cell, for lack of a better word. It used a glass capillary that was permanently installed as opposed to the new technology, which uses disposable, single use cartridges. So it was actually more cumbersome to use, the previous version, than it is now. The commercial system is much easier to use.
We had the technology and we sort of brainstormed about what we could use it for. And one of the first applications was drug resistance development. Taking a cell line and treating it with a chemotherapeutic and mapping out how the cells would become resistant. So what we did was we took K562 cells, which are a leukemic cell line, and we treated with a common chemotherapeutic cisplatin, which is a DNA damaging agent. We treated the cells with low concentrations of the cisplatin and put the cells through the LeviCell, or the previous generation, to separate the live and dead. We would capture the live cells and then reculture them for a number of passages. Each passage we would treat with the drug and eventually, we enriched for cells that were resistant to the drug. Then we used bulk sequencing to look at differences between those cells and the starting population.
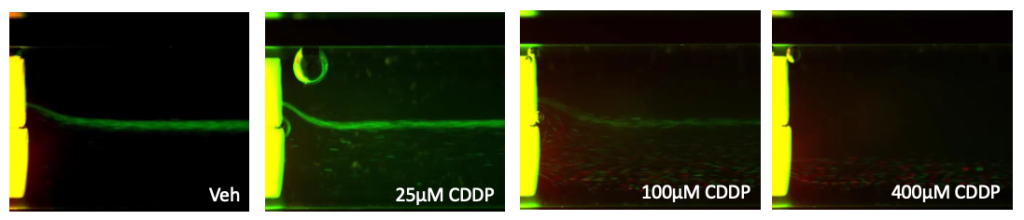
Figure 1. K562 cells were treated with indicated concentrations of cisplatin (CDDP) or Vehicle control (Veh) for 40 hours. Cells were stained with AO/PI as an indicator of live/dead status and subsequently separated by levitation.
The top (viable) and bottom (apoptotic/necrotic/dead) fractions were collected. RNA from collected fractions were extracted, reverse transcribed, prepared with the Ion AmpliSeqTM RNA Apoptosis Panel and sequenced on the Ion GeneStudioTM S5 System.
Could you tell us how you came to work for LevitasBio?
Yeah. I think it’s sort of a funny story. Martin [CEO Martin Pieprzyk] gave me a phone call and he said he’s looking for somebody and do I know anybody? At the time, I wasn’t really looking to change roles, but I listened to what he said. I had been very excited about the new instrument because we had just received the commercial version about a month or two before the phone call. So I thought he just wanted to chat about the new instrument and how we were doing. It became clear that he was interested in bringing me in, and sometimes you just have to take a leap. I believed in the technology. I used it firsthand. I thought it was potentially a game changer, especially for the single cell genomic space, and also for drug development and potential clinical applications in the future.
How did the commercial version of the LeviCell compare to the earlier version you had been working with?
I thought the pre-commercial instrument was easy to use, and the commercial system is orders of magnitude easier to use. You can learn how to use the system efficiently in about 20 minutes. There’s a very steep learning curve on something like a FACS sorter. The LeviCell is a cartridge-based system. You basically put the cartridge in, you load your cells, you press go on the instrument and the instrument does the work for you. There’s really not a lot of guesswork that the operator has to do. And once you’ve used it a couple of times, you are an absolute expert. It’s very easy.
And especially for a lab, like a core facility that would have a lot of different users using the instrument, compared to something like a FACS sorter where basically you need to have a single trained operator that operates the instrument and people just come and use it with that person holding their hand. This instrument doesn’t need any of that. Basically the software wizards you through the steps of operation. There’s no trick to it. It’s very straightforward.
What do you see as the potential of this technology in working with single cell applications, and how does it impact pain points in those research workflows?
Right now, one of the biggest pain points in single cell sequencing and single nuclei sequencing is the quality of the sample at arrival. At the place that’s actually doing the work, what’s the quality of that sample? And so if you’re given a fresh sample that’s never been frozen, the sample quality is pretty good. But that’s really not realistic. Even samples that have come from a different place on campus, extracted at a hospital and moved a quarter mile down the road to the research facility, they’ve been on ice for hours and you start getting cell death, you start getting gene expression changes. And so being able to clean that sample up so you’re only working with the most pristine, most healthy, viable cells to then go into your single cell sequencing work, then suddenly your data is going to be much closer to what the truth is, because we’re ultimately interested in what is going on in vivo, in the body. And when you extract the cells, they’re changing immediately. So the closer we can get to the truth and what’s happening in the real world, the better, right?
Some single cell gene expression protocols, for example, require greater than 80% to 85% viability to guarantee good results. You’re not going to get that from a tissue sample that’s been shipped across the country. Typically, a PBMC sample that comes out of a freezer that’s been stored for years in liquid nitrogen, the viability coming out of that vial could be anywhere from 50 to 75, 80 percent. So at the high end of typical sample viability, you’re at the low end of what’s acceptable for doing single cell work. You could take that sample, put it through the LeviCell, and then about 20 minutes later, you have your sample that’s now at 85, 90, 95 percent viability and your sequencing results are that much better.
Could you speak to how the LeviCell achieves that without introducing additional bias into the system?
The principle that the LeviCell works on is magnetic levitation and inherent buoyancy and magnetic properties of the cells themselves. The cells are resuspended in an inert buffer. We typically use either a PBS- or a cell culture media–based buffer that has an agent in it that interacts with the magnet and causes a magnetic field across the solution so the cells then levitate based on their inherent buoyancy and magnetic properties. So we’re able to separate cells without otherwise changing the cell at all. We don’t need to label it with an antibody or dye it in any way or somehow perturb the cell to then capture it. The cell is straight from its source and into the system, just resuspended in this buffer. The workflow is quite quick. And then at the end, you have this cell that’s ready to go into your downstream application, whether it’s sequencing or cell culture or any sort of functional analysis.